Understanding DNA replication dynamics in E. coli
Meet the Experts

Christian Rudolph, Principal Investigator/Reader, Brunel University London
Email: [email protected]
Tel: +44 (0)1895 265372
Despite incredible advances in genetic research, gaps in our knowledge still remain, even in a bacterium like E. coli, which has been a model organism for many decades.
At Brunel University London, Dr Christian Rudolph seeks to better understand DNA replication and chromosome dynamics in E. coli, with a particular focus on the termination stage. He explains his research and how upgrading his microscope’s light source from metal halide to the CoolLED pE-4000 Illumination System has enhanced his time lapse studies.
Can you tell us about your research?
I’ve been a PI at Brunel University for almost 10 years, where I run my own research group. My long-standing research interest is genomic stability. The genome of a bacterium such as E. coli consists of just under five million nucleotides, and humans have a thousand times more. But still, every time cells replicate their genome, the resulting daughter cells have a good chance of being genetically identical. We want to understand how, in the case of E. coli cells, hundreds of copies of the genome can be generated without any errors. Understanding these fundamental aspects of quality control and accuracy is an important aspect in understanding tumourigenesis on one hand, but will also help with the development of novel antibacterial compounds.
Currently our main focus is to develop a detailed understanding of what happens at the end of genomic replication in living cells. For DNA replication to be completed, two complex and fast-moving replication complexes have to come together. As part of this process every single base pair in between the replication fork complexes has to be duplicated with high accuracy in order to produce two complete chromosomal copies. These final steps are surprisingly complex, and they have been a focus of my research for the past decade. It was simply assumed the forks just come together, but we have learned that many things can go wrong in bacteria – and we have only just started to look at what happens in eukaryotic cells. E. coli is an ideal model for this work, largely because it has served as a model for many decades and therefore our understanding is much better. But in addition, E. coli cells only have a single chromosome, exactly two replication fork complexes and only one fusion event – and we know exactly where fusion takes place. In yeast cells, for example, the presence of hundreds of origins of replication results in hundreds of fork fusion events, and so it gets complex very quickly.
We’re essentially working towards an improved model of how cells work, which is part of what drives my research. By providing new ideas and improved models in bacteria, we have already seen other research groups around the world pick this up and start work in eukaryotic cells.
Which research techniques do you use?
On one hand, we are interested in understanding where the terminus region of the chromosome is in the cell, which is currently achieved using a fluorescent repressor-operator system (FROS). Alongside this we use a lot of fluorescently labelled proteins, such as the replication machinery. In some cells we can actually visualise both the replication fork and termination area simultaneously. We can see the complex approach and understand, for example, how and when that specific area of the chromosome is duplicated or segregated. In terms of termination, there is still plenty to understand.
Another project in collaboration with Dr Ed Bolt at the University of Nottingham is to understand what CRISPR-Cas systems are precisely doing in bacterial cells. As a result of this work we are currently looking into whether conventional FROS approaches can be replaced by CRISPR-Cas-based approaches, allowing the easy tagging of specific chromosomal areas. In theory it is quite simple: a guide RNA for a specific area such as the termination area of the chromosome is generated and expressed in cells which carry a fluorescent Cas protein that will no longer cleave DNA. The tagged Cas protein will then bind to the chromosomal area defined by the guide RNA. This works well for example in plant cells, but I do not think anybody has tried this approach in bacteria yet.
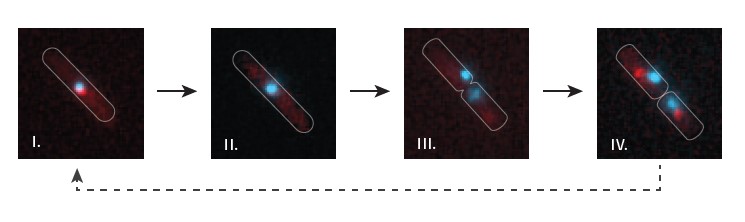
How has upgrading your fluorescence light source helped your work?
We rely a lot on time lapse microscopy, and if we acquire an image every five or 10 minutes over a longer period of time, photobleaching of our fluorophores becomes a significant problem. With the metal halide system, we did not play around with the illumination much because swapping neutral density filters in and out makes the process quite tedious. But of course, now with the pE-4000 we have a simple intensity slider. I sat down just for a bit of fun, essentially, and ran a time lapse to play around with the new light source, and it was incredibly easy.
Another problem is that different fluorophores react differently to exposure. For some fluorophores it can be beneficial to expose your sample for twice as long, but at a 50% reduced intensity. In contrast, for other fluorophores the total time of exposure is more important. It is important to determine the characteristics of the fluorophores in use. In my lab we are only using conventional widefield illumination, but we visualise complexes where the copy number is very low. For example, in active replication fork complexes there are only three fluorescent copies per complex. We can visualise this with our system, but time lapse microscopy was not really feasible using metal halide illumination. The illumination control of the pE-4000 has greatly improved our time lapse capabilities. We have spent some time experimenting with longer exposures and reduced intensities and that helps, enabling us to acquire more frames from a time lapse experiment.
Are there any other differences compared with the metal halide light source?
Firstly, you do not need to worry about running time with the pE-4000, and even if you do accidently leave it on overnight, it’s not the end of the world.
Initially, one thing that I thought I needed was a TTL connection with the camera, although for the type of work we do, the extra speed is not required, especially as the system is much faster than our previous metal halide system. From my perspective the pE-4000 has improved our system in many ways; it is one of the best upgrades we have done recently, and I would never go back.
Learn more about this research
Find out about the CoolLED pE-4000 Illumination System
Thank you to www.digitalpixel.co.uk for supplying the microscope upgrade for the Rudolph Lab
